Statistics or biology: the zero-inflation controversy about scRNA-seq data
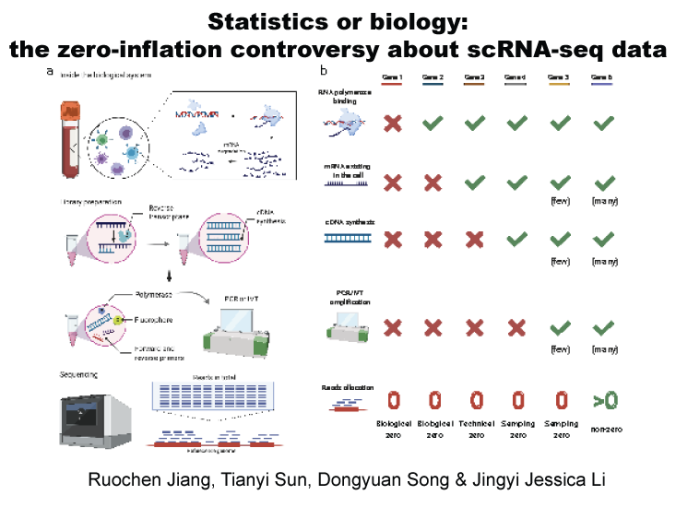
Single-cell RNA sequencing (scRNA-seq) technologies have revolutionized biomedical sciences by enabling genome-wide profiling of gene expression levels at an unprecedented single-cell resolution. A distinct characteristic of scRNA-seq data is the vast proportion of zeros unseen in bulk RNA-seq data. Researchers view these zeros differently: some regard zeros as biological signals representing no or low gene expression, while others regard zeros as false signals or missing data to be corrected. As a result, the scRNA-seq field faces much controversy regarding how to handle zeros in data analysis.
In this paper, we first discuss the sources of biological and non-biological zeros in scRNA-seq data. Second, we evaluate the impacts of non-biological zeros on cell clustering and differential gene expression analysis. Third, we introduce five mechanisms of adding non-biological zeros in computational benchmarking. Fourth, we summarize the advantages, disadvantages, and suitable uses of three input data types: observed counts, imputed counts, and binarized counts. Fifth, we benchmark the performance of the three input data types in cell clustering, dimension reduction, and differential gene expression analysis. Finally, we discuss the open questions regarding non-biological zeros and the importance of transparent analysis.
